Amplicon-EZ Frequently Asked Questions
Review our Amplicon-EZ Best Practices for sample submission
Sample Preparation
What types of samples can be submitted for Amplicon-EZ?
Purified PCR products 150-500 bp, with or without partial Illumina® adapter sequences, at least 500 ng double-stranded DNA total, normalized to a concentration of 20 ng/µL, submitted in microcentrifuge tubes. For optimal results, we recommend submitting samples that produce only one band on a gel.
Does GENEWIZ perform QC on my Amplicon-EZ samples prior to library preparation?
No, we do not perform sample QC in an effort to provide the fastest and most cost-effective service possible. Please measure the concentration of your samples (preferably using a dsDNA quantification assay such as Qubit or PicoGreen) and normalize to 20 ng/µL prior to submission.
Does the starting material need to be normalized to 20 ng/µL?
Yes, GENEWIZ does not adjust the concentration prior to library preparation. Instead, we use equal volumes for each sample. For optimal results, please adjust your sample concentrations to 20 ng/µL prior to submission. We recommend using a dsDNA quantification assay such as Qubit or PicoGreen.
Note that the NanoDrop cannot distinguish between dsDNA and ssDNA (oligos and dNTPs) and may cause you to overestimate the amount of dsDNA in your samples.
Can I submit products that are not in the 150-500 bp size range for Amplicon-EZ?
No, only PCR products 150-500 bp are accepted for Amplicon-EZ. However, you can submit samples outside this range using our other amplicon sequencing services. For Amplicon-EZ, we perform 2x250 bp sequencing without fragmenting the amplicons.
What are the partial Illumina® adapter sequences?
For the forward sequencing read: 5’-ACACTCTTTCCCTACACGACGCTCTTCCGATCT-3’
For the reverse sequencing read: 5’-GACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3’
Amplicons with the partial adapters should be constructed as follows:

How do I add partial adapters to my amplicons?
We suggest designing PCR primers with the adapter sequences added to the 5' end of your primers.
How should I purify my amplicons?
We do not currently have a preferred or recommended method for sample purification. We have successfully processed samples purified via DNA-binding beads/columns, enzymatic cleanup and gel purification.
What happens if my samples do not meet your starting material requirements?
Sample Submission
When can I submit samples?
Samples can be submitted at any time; however, they are processed in cycles starting on Wednesdays. Samples must be received by 3 pm ET Wednesday at our NJ facility to be eligible for that week’s cycle. Projects with incomplete information may be placed in the next sequencing cycle. Results are delivered 8 business days after the initial day of processing (on the second Monday).
What are the sample submission guidelines?
Sample Type: Purified PCR Products
Size: 150-500 bp (For optimal results, we recommend samples that produce only one band on a gel.)
Minimum Amount: 500 ng (We recommend using a dsDNA quantification assay such as Qubit or PicoGreen.)
Concentration: Normalized to 20 ng/µL
Purity (A260/280): 1.8-2.0
Buffer: Water, EB, or low TE (<0.1 mM EDTA)
(For more details, see the Sample Preparation section)
How should I prepare the tubes?
Prepare samples in microcentrifuge tubes. Label each tube clearly with the sample name, as it appears on the order form. See example below.
How can I send my samples to GENEWIZ?
Option #1: Submit samples into a local GENEWIZ dropbox which are conveniently located throughout the US. To locate a dropbox near you, please submit an inquiry. Place your samples and order receipt together in a Ziploc bag at room temperature.
Option #2: Ship samples directly to our facility. Use the shipping address listed on your order receipt. You can ship at room temperature or on blue/dry ice.
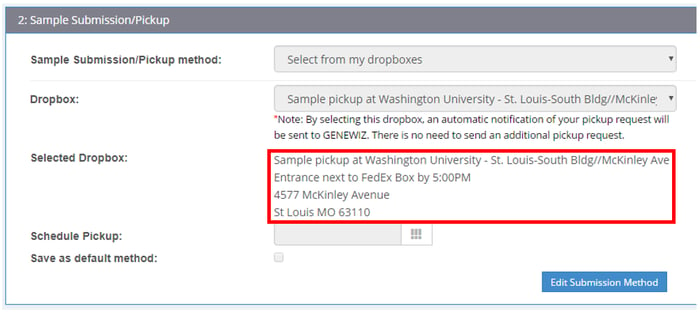
What are the cutoff times for my dropbox pickup?
Consult the Order Summary page to find out the daily cutoff times for your selected dropbox (see example below). Samples must be received at our NJ facility by Wednesday to be included in that week's processing cycle. For US customers on the west coast, please submit samples by Monday to meet this deadline. For all other US customers, please submit by Tuesday. If you miss the dropbox deadline, feel free to ship samples directly to us.
Order & Processing
How do I order Amplicon-EZ?
You can order Amplicon-EZ in minutes without a quote. Check out our Ordering Guide to view the six easy steps or watch the 2-minute video below.
How can I monitor the progress of my Amplicon-EZ project?
The estimated delivery date for your order can be viewed anytime through your GENEWIZ account. Visit the Order Summary page. Because of the fast turnaround time of the service, we do not provide detailed project updates (i.e. timestamps for QC, library preparation, sequencing, and data analysis).
What is the turnaround time?
Samples are processed on weekly cycles starting on Wednesday. Results are available within 8 business days from the initial day of processing.
Results
How much data can I expect per sample?
GENEWIZ typically provides ~50,000 reads per sample.
Will raw data be delivered to me?
Yes, all raw data (i.e. FASTQ files) will be delivered upon project completion.
How do I download my data?
Once the data analysis is complete, if applicable, a member of our bioinformatics team will reach out to you with a link to download your data from our sFTP server. Login credentials and instructions on how to retrieve your data will be included in the data delivery email. Please feel free to contact us should you have trouble retrieving your data. You can also refer to GENEWIZ sFTP Data Download Guide for step-by-step instructions and troubleshooting tips.
Will my Amplicon-EZ data be secure at GENEWIZ?
We take data security very seriously and we make every attempt to keep your data private and protected. The data transfer we offer is secure; however, should you desire an alternative delivery method, we are happy to work with you.
What type of analysis will I receive?
The type of analysis depends on amplicon size and the information provided in "Step 3: Data Analysis" on the order form. See the flow chart below to determine which analysis is available for your samples.
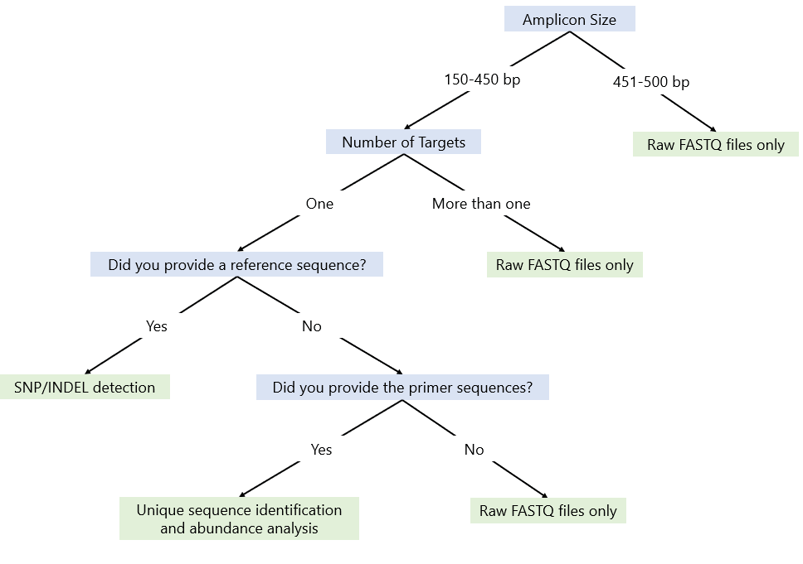
What does the SNP/INDEL detection analysis include?
This analysis generates an interactive HTML-based report that includes variant detection (SNPs and INDELs up to 20 bp) as well as genotyping information (homozygous wild type, heterozygous, and homozygous mutant). Unique sequences will also be quantified. Each sample will be represented individually in a user-friendly interface.
What does the unique sequence identification and abundance analysis include?
This analysis generates results that include a list of unique sequences up to 450 bp and their relative abundance in the sequencing data.
Do you offer analysis for projects that have multiple primer pairs and/or reference sequences per sample?
This is considered a custom request. Please submit an inquiry for further information
Is there additional bioinformatics available?
All custom bioinformatic analysis requests are taken on a case-by-case basis after a technical consultation with one of our NGS study managers. Please note that any customization outside our standard deliverables will incur additional charges and time.
What are some examples of additional bioinformatics?
Examples include de-multiplexing based on in-line barcodes, sample-to-sample comparisons, de-multiplexing of a large number of amplicons, and heavy/light chain analysis.
Troubleshooting
What if my sequencing run produces fewer than 50,000 reads?
We strive to achieve >40,000 reads per sample. If 10,000-40,000 reads are generated, we can provide additional sequencing to "top off" the data upon request.
What if my sequencing run yielded fewer than 10,000 reads?
Based on historical troubleshooting, this result indicates a potential sample-related issue. We would highly recommend reviewing our Amplicon-EZ Best Practices for more details and preparing new samples.
My sample showed multiple bands on a gel, but I don't see all PCR products represented in the sequencing results. Why?
Our Amplicon-EZ workflow is optimized for PCR products represented by a single band on a gel within the 150-500 bp size range. You can submit samples with multiple bands; however, we cannot guarantee successful library creation or sequencing results for these types of samples. Since fragment size influences sequencing efficiency, samples with PCR products of varying sizes may not show uniform coverage.
What if my results do not match my expectations or hypothesis?
We cannot guarantee the actual results of the sequencing data. If you feel that a processing error may have occurred, please contact us and we can investigate on a case-by-case basis.
Have a specific question?
Contact Us | Phone 1-877-GENEWIZ (436-3949), ext. 1