Sample Preparation
1. What sample types should I prepare for PCR-EZ?
- Clonal, linear, purified, double-stranded DNA (dsDNA) PCR products normalized to a concentration of 50 ng/µL in a minimum volume of 10 µL submitted in microcentrifuge tubes, sealed 96-well plates, or capped PCR strip tubes.
- Clonal, linear, unpurified, double-stranded DNA (dsDNA) PCR products in a minimum volume of 10 µL submitted in microcentrifuge tubes, sealed 96-well plates, or capped PCR strip tubes.
- Single-stranded DNA (ssDNA) are not supported at this time and results are not guaranteed.
2. Does the starting material need to be normalized to 50 ng/µL?
- For purified samples: Yes. We do not adjust the concentration prior to library preparation. Instead, we use equal volumes for each sample. For optimal results, please adjust your sample concentrations to 50 ng/µL prior to submission. We recommend using a dsDNA quantification assay such as QubitTM or PicoGreenTM.
Note: NanoDropTM cannot distinguish between dsDNA and ssDNA (oligos and dNTPs) and may cause you to overestimate the amount of dsDNA in your samples. - For unpurified samples, we recommend providing a gel image to help us estimate DNA concentration and optimize the dilution after enzymatic cleanup.
3. What is the size range of PCR amplicons that I can submit for sequencing?
PCR amplicons 500 bp to 25 kb can be sequenced using this workflow. If you are sequencing larger than 25 kb, please submit an inquiry for a technical consultation.
4. Do I need to send primers with my samples?
No, primers are not needed. Samples are sequenced without primers; therefore, please do not send or mix primers with your sample submission.
5. Can a mixture of PCR amplicons be sequenced using this service?
No, this service is intended for a clonal population of PCR amplicons. See Troubleshooting for more details.
6. What happens if my samples do not meet your starting material requirements?
Please submit an inquiry and someone will be in touch to discuss options.
Sample Submission
1. When can I submit my samples?
Samples may be submitted at any time. Samples must be received in the morning at our facility to be eligible for that day's cycle. Projects with incomplete information may be placed in the next sequencing cycle. Results are delivered the same day in select regions or in as fast as 1 business day.
2. What are the sample submission guidelines for PCR-EZ?
- Purified Linear PCR:
- Sample Type: Purified Amplicon DNA
- Size: 500 bp - 25 kb; if larger than 25kb, please submit an inquiry for a technical consultation
- Minimum Amount: 500 ng (we recommend using a dsDNA quantification assay such as QubitTM or PicoGreenTM)
- Concentration: Normalized to 50 ng/µL
- Purity (A260/280): 1.8-2.0
- Buffer: Water, EB, or low TE (<0.1 mM EDTA)
-
Unpurified Linear PCR:
- Sample Type: Unpurified Amplicon DNA
- Size: 500 bp - 25 kb; if larger than 25kb, please submit an inquiry for a technical consultation
- We recommend providing a gel image to help us estimate DNA concentration and optimize the dilution after enzymatic cleanup
- Buffer: Water, EB, or low TE (<0.1 mM EDTA)
3. How should I prepare the tubes/plates?
Tubes for <48 Samples
- If you are submitting <48 samples, please use 8-strip, 0.2 ml PCR tubes and caps. You can cut off empty tubes and use them next time.
- Our online ordering system will assign tube ID codes using your initials and the sample number (see figure below). Please write these codes on the sides of your tubes using indelible marker.
- Label the side of your tubes with YOUR initials and sample number. These should match your order receipt.
![]()
Plates for >48 Samples
- If you are submitting 48 or more samples, we recommend using a 96-well, semi-skirted PCR plate. Cap the wells with 8-strip caps. These are usually ordered separately from the plates. Be sure that the caps seal tightly! The semi-skirted plate helps to prevent the plate from bending in transit, resulting in fewer loose caps. Please ship the plate with cushioning to avoid potential damage.
- Please contact us for help finding a vendor if your laboratory does not have any 96-well PCR plates or matching strip caps.
- Arrange your samples vertically (in columns) as shown below:
.![]()
4. How can I send my samples?
Please send your samples with a copy of your order receipt to us using one of the following options.
Option #1: Submit samples into a local dropbox at room temperature. We have an extensive network of convenient dropboxes located throughout the US. To find a dropbox near you, please submit an inquiry. Place the samples and order receipt together in a Ziploc bag.
Option #2: Ship samples directly to our facility using the shipping address listed on your order receipt. You can ship at room temperature or on blue/dry ice.
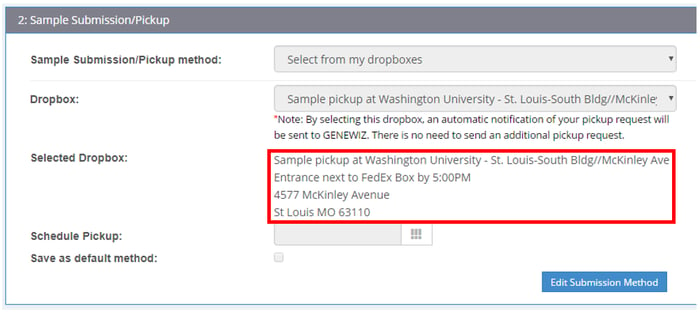
5. What are the cutoff times for my dropbox pickup?
When submitting an order in our customer portal, the final step will locate the nearest dropboxes to your institution. You can then choose the dropbox where you will leave your sample for PCR-EZ. During checkout, consult the Order Summary page to find out the daily cutoff times for your selected dropbox (see example below).
If you miss the dropbox deadline, feel free to ship samples directly to us.
Ordering & Processing
1. Why should I choose PCR-EZ?

*If you are sequencing larger than 25 kb amplicons, please submit an inquiry to https://web.genewiz.com/ngs-inquiry for a technical consultation.
2. How do I order PCR-EZ?
You can order PCR-EZ in minutes without a quote. Visit genewiz.com and login to your online account to place an order.
3. How much sequencing coverage is expected per amplicon?
Coverage is highly dependent on sample quality as determined by end-user prior to sample submission and therefore, cannot be guaranteed for this service.
4. How can I monitor the progress of my PCR-EZ project?
The estimated delivery date for your order can be viewed anytime through your online account, on the Order Summary page. Due to the fast turnaround time of this service, we do not provide detailed project updates (i.e. timestamps for QC, library preparation, sequencing, and data analysis).
5. What is the turnaround time?
Samples are processed every weekday. Results are available the same day in select regions or in as fast as 8:00 AM the next day after sample receipt.
.
6. Do you perform QC on my PCR-EZ samples prior to library preparation?
No, we do not perform sample QC. This enables us to provide rapid and cost-effective service for amplicon sequencing. Please measure the concentration of your purified samples (preferably using a dsDNA quantification assay such as QubitTM or PicoGreenTM) and normalize to 50 ng/µL prior to submission.
Results
1. What data output will I receive for PCR-EZ?
For clonal PCR products: an interactive data report, consensus sequence of amplicon in fasta format, raw reads in fastq files, and a sequence variant report.

2. How do I download my data?
Once data analysis is complete, your data will be delivered electronically by sFTP link, email, or through your online account. If applicable, login credentials and instructions on how to retrieve your data will be included in the data delivery email sent to you. Please feel free to contact us should you have trouble retrieving your data. You may also refer to our sFTP Data Download Guide for step-by-step instructions and troubleshooting tips.
3. Will my PCR-EZ data be secure?
Yes, we take data security very seriously and make every attempt to keep your data private and protected. The data transfer we offer is secure; however, should you desire an alternative delivery method, we are happy to work with you.
4. Is there additional bioinformatics available?
All custom bioinformatic analysis requests are taken on a case-by-case basis. Please submit an inquiry for a technical consultation. Please note that any customization outside our standard deliverables will incur additional charges and time.
Troubleshooting
1. What if my amplicon samples fail to sequence or fail to generate a consensus sequence?
It is likely that sequencing failure (i.e. failure of the sample to produce consensus sequence with at least 10x coverage or less) may be due to samples not prepared at the required DNA concentration, containing a mixed plasmid population, or degraded/fragmented DNA.
To achieve optimal sequencing results, please follow our recommended sample submission guidelines.

2. What if my results do not match my expectations or hypothesis?
We cannot guarantee the results of the sequencing data. If you feel that a processing error may have occurred, please contact us and we will investigate on a case-by-case basis.