Precision Antibody Discovery Frequently Asked Questions
General Questions
What is Precision Antibody Discovery?
GENEWIZ Multiomics & Synthesis Solutions from Azenta Life Sciences offers Precision Antibody Discovery, an innovative, end-to-end antibody screening solution that guides your discovery program to uncover unique antibody leads with more favorable development characteristics. Utilizing next generation sequencing (NGS) of your in vivo samples (i.e., B cells, PBMCs) or in vitro libraries (i.e., phage display) and our bioinformatic platform, AbXtract™ (powered by OpenEye and developed by Specifica), we rapidly identify and produce antibody sequences with the most promising biophysical profiles. Following upstream analysis, recombinant antibodies are then generated via gene synthesis for expression and purification.
Our Precision Antibody Discovery workflow utilizing AbXtract is illustrated below.
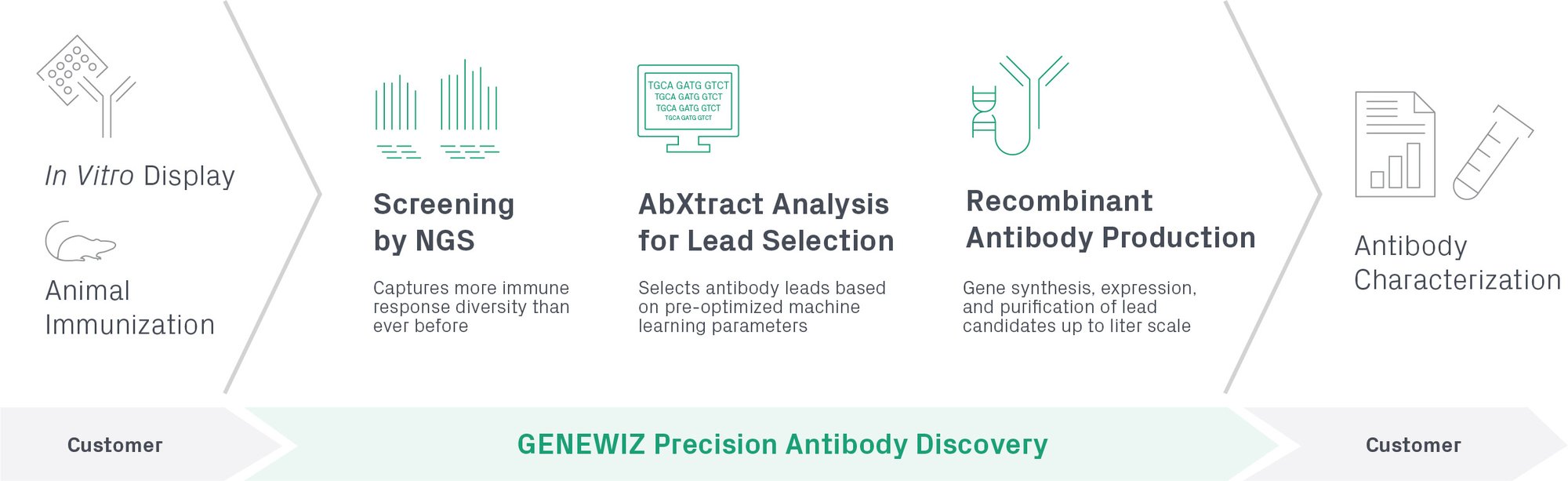
What is AbXtract?
Using the power of machine learning, AbXtract is a novel bioinformatic platform that enables high-throughput screening of your library to generate 5 – 50x more antibody leads than traditional low-throughput approaches. Utilizing AbXtract can improve the success of your antibody development program with a greater number of leads and access to rare clones for antibody discovery.
How does AbXtract work?
The AbXtract workflow is based on 3 main steps:
1. Data Upload
Quality filtering of the input sequence data is followed by sequence annotation to identify regions of interest (like CDRs) and extra features:
- Annotate to identify CDRs and framework regions of interest (IMGT, Kabat, Chothia, or custom annotation)
- Produce annotated records ideally suited for antibody discovery
2. Cluster Antibody
Relative abundance and enrichment based on CDRs is calculated. The module then identifies abundant and rare clusters with unsupervised machine learning (ML) based on sequence features.
Antibodies in the same cluster are expected to have the same paratope and hence bind the same epitope. Exploring additional antibodies in the same cluster can yield antibodies with the same biological activity but spanning a range of affinities with reduced sequence liabilities. Antibodies in different clusters are more likely to bind different epitopes but can also bind overlapping epitopes.
3. Optimal Antibody Leads
The platform prioritizes leads based on favorable NGS metrics (i.e., optimized for diversity, abundance, enrichment, reduced redundancy, and sequence liabilities).
Is GENEWIZ Precision Antibody Discovery compatible with either in vitro display screening or in vivo immunization?
Yes, the analysis platform can support both antibody discovery approaches.
Sample Preparation & Submission
What sample types do you accept to initiate antibody discovery screening?
For in vivo samples, we accept total RNA or biological material for RNA extraction.
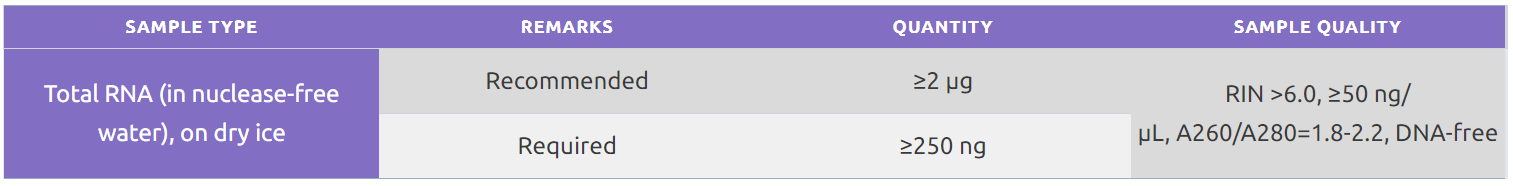
For RNA extraction, we recommend a frozen cell pellet of 10^6 cells with a minimum submission of 10^5 cells. For other types of starting material, please submit an inquiry.
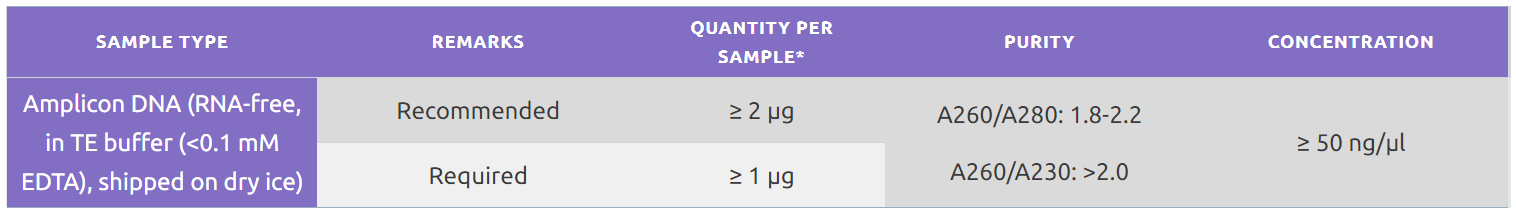
- For phage display libraries, we can accept an amplicon generated from the heavy and light chain.
Ordering & Processing
How can I get paired heavy and light chain data?
Physical pairing of the VH/VL can be achieved with long-read sequencing using PacBio™ technology. Short-read sequencing can provide unpaired VH/VL data. Light and heavy chain can be sequenced separately with short reads. In that case, we suggest only running AbXtract on the heavy chain. We can then pair it with your VL of choice from a select list to proceed with antibody production.
How much time does it take to synthesize DNA for antibodies?
We have the fastest turnaround time on the market for antibody DNA synthesis, starting at 6-8 business days for up to 1,500 bp. There is no additional turnaround time for cloning into custom vectors, including bicistronic vectors. We can clone into any vector provided; synthesis is compatible with all antibody formats.
What is the ordering/project management process for GENEWIZ Precision Antibody Discovery?
First, we will provide a consultation to review the objectives of your project to ensure that goals are outlined and agreed upon. After the consultation, you will receive a quote capturing details of your project, pricing, and turnaround time. Once the project starts, you will receive periodic updates throughout the NGS, data analysis, gene synthesis, and antibody production steps.
Results
Will I receive a report?
Yes, a full sequence of the synthesized leads and a report is provided. The report includes:
- General stats for CDR3s chains and clusters across all samples
- Top 100 clusters, ranked by group-wise abundance
- Cumulative sequence liabilities per chain in CDRs
Who has access to my data and how is my data stored and secured?
Only trained Azenta scientists can access the data in AbXtract. We use advanced encryption protocols to ensure the safety and integrity of your data. We use the AWS Transfer Family service which is GDPR compliant.
All our employees are well-trained on cybersecurity and mitigation tactics and tested regularly to ensure compliance. We have external monitoring and testing in place from independent third parties to ensure that we have no blind spots.
What deliverables will I receive with GENEWIZ Precision Antibody Discovery?
- FASTQ files
- AbXtract report
- Purified antibodies
- Antibody QC report including SDS-PAGE analysis
Have a specific question?
Contact Us | Phone 1-877-GENEWIZ (436-3949), ext. 1