Jump to Topic
Assay FAQ:
GENEWIZ FAQ:
What Sequencing Only services does GENEWIZ offer?
We offer high-throughput next generation sequencing (NGS) on pre-made NGS libraries using Illumina® platforms.
What type of pre-made libraries can I submit?
You can submit any Illumina®-compatible library for sequencing. If the library preparation used Illumina® adapters during ligation, these will be successfully sequenced using GENEWIZ’s Sequencing Only service.
What is the size range of pre-made libraries that I can submit?
You can submit libraries up to 600 bp.
Do you offer next generation sequencing of pre-made libraries on platforms other than Illumina®?
Yes, we offer next generation sequencing of pre-made NGS libraries on other NGS platforms, including long-read platforms. Please contact the team by submitting an inquiry.
Can you spike PhiX into my sequencing run?
Yes, please instruct our team what percentage of PhiX spike-in to use, and we’ll accommodate the request free of charge. See the following recommendations of PhiX spike-in:
- ~20% taxon identification (ITS/16S/18S) and WGBS
- ~30% amplicon and small RNA libraries
- ~5%-10% low diversity libraries
Please indicate that you require data analysis upon submitting your instant quote.
![]()
Do you accept libraries with custom sequencing or index primers?
Yes, please indicate that your libraries have custom sequencing or index primers upon submitting your instant quote.

What sequencing configurations are available?
There are several Illumina® sequencing configurations available including 2x150 bp, 2x250 bp, 2x300 bp, and 10x sequencing.
GENEWIZ has a variety of sequencers, offering different amounts of data output:
- 2x150 bp configuration: MiSeq flow cell (4.5 Gb), NovaSeq X Plus 10B flow cell (3,000 Gb), NovaSeq X Plus 25B lane (950 Gb), NovaSeq X Plus 25B flow cell (7,600 Gb)
- 2x250 bp configuration: MiSeq flow cell (7.5 Gb), NovaSeq 6000 SP flow cell (325-400 Gb)
- 2x300 bp configuration: MiSeq flow cell (13 Gb)
- 10x sequencing only: NovaSeq X Plus 25B lane (950 Gb), NovaSeq X Plus 25B flow cell (7,600 Gb)
What is a flow cell?
A flow cell is a glass or polymer slide used in NGS platforms (like Illumina®) where DNA fragments are immobilized, amplified, and sequenced. It contains multiple lanes or channels that can be loaded with different DNA libraries.
What is a dedicated lane/flow cell?
A dedicated lane/flow cell means ordering and utilizing the entire lane of a flow cell or the entire flow cell. The lane/flow cell is dedicated to your individual project, maximizing throughput and minimizing cross-contamination risk.

What is a partial lane/flow cell?
A partial lane/flow cell refers to using only a subset of the lane of a flow cell. The remaining lanes may be used for other projects or pooled with other users’ samples. This is often called lane sharing or multiplexing across lanes. This option is only available for 2x150 bp configuration.

There are several data packages available: 105 Gb, 375 Gb. The data package will be assigned based on the total number of libraries, amount of data per library, and amount of PhiX spike-in required.
When would you use a dedicated lane/flow cell?
- Large-scale projects needing high read depth
- Clinical or regulatory-grade sequencing where contamination must be avoided
- Require sequencing configuration outside of 2x150 bp (i.e. 2x250 bp, 2x300 bp)
- Require specific Illumina sequencing platform to keep continuity for publication purposes
When is a partial lane/flow cell more appropriate?
- Smaller projects with fewer samples needing less than 950 Gb of data output
- Cost-conscious sequencing where full lane/flow cell capacity isn’t needed
- Sequencing configuration of 2x150 bp
Is there a risk of sample mix-up in partial flow cells?
The risk is minimal, but possible. GENEWIZ has an optimized and efficient process of pooling samples from different projects to ensure the highest quality and chain of custody.
Do both options use the same sequencing chemistry?
Yes, the sequencing chemistry remains the same. The difference lies in how the lane/flow cell is allocated and utilized.
What is the price for Sequencing Only?
Please submit a quote request to receive accurate pricing information, as the cost depends on the details of the project. Please submit an inquiry to inquire about your specific project needs.
Partial lane/flow cell runs are less total money because you pay for a smaller data package. Dedicated lane/flow cell runs are more expensive but more cost-effective by cost per Gb.
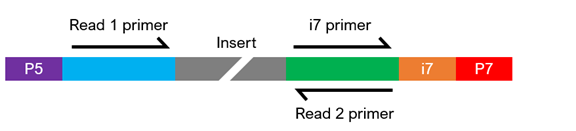
What does it mean to be a standard Illumina-compatible library?
Illumina®-compatible libraries require four major components in their adapter sequences, found at the ends of each fragment, for paired-end sequencing:
- Flow cell binding sites. Known as P5 or P7, they facilitate clustering of the DNA strands on the flow cell.
- Sequencing primer binding sites. Labeled as Read 1 and Read 2, they are necessary to initiate sequencing at both ends of the fragment.
- Index sequences. Often called i5 and i7, they act as barcodes to allow multiplexing (pooling) of samples. Libraries can be either single index (having one index sequencing) or dual index (having two index sequences), and each library to be pooled requires at least one unique index.
- Index primer binding site(s). These are required to read the index sequences.
The composition of the adapter sequences depends on whether the library is single or dual indexed (see figure below). If any of the components are missing, heavily modified, or in the wrong configuration, sequencing may fail. If you have any questions regarding your library configuration, please contact us.
Single-Indexed Sequencing
Dual-Indexed Sequencing
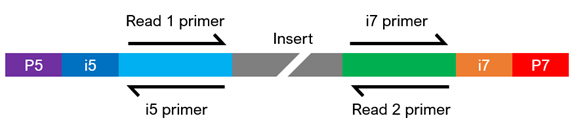
Please indicate what type of indexed sequences (index primer binding sites) your pre-made libraries contain upon submitting your instant quote.
![]()

What index sizes can be accepted?
GENEWIZ can accept sequencing libraries with index configurations of 6 bp (most single index applications), 8 bp (most dual index applications), 10 bp, and 16 bp. Other custom index options are available. Please submit an inquiry for more details.
Can GENEWIZ de-multiplex my data?
Yes, we will provide the de-multiplexing step free of charge. We ask that you provide the index sequences in the Sample Submission Form prior to your sequencing run.
Can GENEWIZ perform de-multiplexing of “in-line” barcodes in sequencing reads?
Yes, we offer de-multiplexing of in-line barcodes in sequencing reads (not index reads). Additional charges may apply.
Please indicate that you require de-multiplexing of “in-line” barcodes upon submitting your instant quote.


Can you perform data analysis for my Sequencing Only project?
Yes, we offer a variety of data analysis services across all applications including whole genome sequencing, RNA sequencing, single-cell sequencing, exome sequencing, epigenomics, metagenomics, immunogenomics, and proteomics.
Please indicate that you require data analysis upon submitting your instant quote.


How do I contact GENEWIZ for a technical consultation about my project?
The GENEWIZ NGS team is composed of Ph.D. scientists who can help you optimize your project design and provide consultation. You can contact the team by submitting an inquiry.
Does GENEWIZ guarantee turnaround?
Our team prioritizes fast turnaround times and provides a timeline based on first-pass processing. The turnaround time listed within the quote is inclusive of all steps quoted, unless otherwise noted. If repeat processing is required, the turnaround may be subject to change and will be proactively communicated to the client.
If the scope of a project changes after project initiation or if sample or project clarification is required after sample receipt, GENEWIZ may reassess the turnaround time based on the subsequent communications and modifications (if applicable).
Does GENEWIZ perform sample QC and what methods are used?
For most of the services, an initial sample QC is included within the fee of the project for all samples that will proceed to the next processing stage (e.g. library preparation or sequencing) unless otherwise noted within the quotation. Resubmissions or additional, optional samples may incur a nominal fee for QC.
Initial sample QC can include:
- Assessing RNA concentration and integrity
- Assessing DNA concentration and DNA size (for select projects)
- Assessing premade library size and concentration
- Assessing cryopreserved cells by cell count and viability
What sequencing instruments will be used for my project?
The sequencing platform will be listed within the Service Description of the quotation.
Unless specifically noted in the quotation, GENEWIZ reserves the right to choose between equivalent instruments depending on the target read depth and configuration requested. If a specific instrument is required, please add special comments in your quote/order and notify the GENEWIZ NGS team prior to project initiation.
You can contact the GENEWIZ NGS team at:
- US: NGS@azenta.com
- EU & UK: NGS.Europe@azenta.com
What is GENEWIZ’s data analysis guarantee?
Data analysis is performed to the best of our ability and results are not guaranteed for samples or libraries not passing QC or do not fit the analysis pipeline’s criteria.
Can you recommend the best data delivery option?
Yes, we can recommend the best data delivery option based on the platform and project details. Options include:
- Secure File Transfer Protocol (sFTP) - additional charges may apply
- Customer Cloud Account - AWS, Microsoft Azure, Google Cloud
- Hard Drive - additional charges may apply
Delivery to most customer cloud solutions is free of charge. If you have any questions, please submit an inquiry.
How will my data be delivered?
By default, results are sent via a secure File Transfer Protocol (sFTP). Please refer to our sFTP Data Download Guide for instructions on how to download your data and troubleshooting tips. Additional charges may be applicable for large data transfers.
Please refer to the delivery email sent from GENEWIZ for detailed information on the transfer and consult with your IT department to ensure compliance with your institution’s policies.
Will my data be secure at GENEWIZ?
We take data security very seriously and we make every attempt to keep your data private and protected. The data transfer we offer is secure; however, should you desire an alternative delivery method, we are happy to work with you.
How long does GENEWIZ store samples?
We hold any remaining samples for up to 3 months after project completion. Clinical samples processed in our Regulatory-Environment or CLIA-licensed laboratory may have longer sample storage timelines. Please contact us if you would like samples retained for a longer period or shipped back to you.
How long does GENEWIZ store data?
GENEWIZ offers raw data storage (i.e. FASTQ for Illumina, BAM for PacBio, POD5 for Oxford Nanopore) for up to 6 months after project completion. Data generated for samples processed in the Regulatory-Environment or CLIA-licensed laboratory may have longer data retention timelines. Please contact us if you would like the data to be retained for a longer period.
How should I prepare and send my samples?
View our Sample Submission Guidelines for instructions on preparing and sending samples. Organize your samples in tubes or plates following the order indicated on the sample submission form.
- Tubes: Prepare samples in clearly labeled and well-organized 1.5 mL flip-cap microcentrifuge tubes. Please avoid using Parafilm to seal the tubes.
![]()
Unless otherwise instructed, GENEWIZ reserves the right to combine multiple vials of the same sample for extraction and/or library preparation.
- Plates: For projects with 16 or more samples, prepare samples in clearly labeled, securely sealed 96-well full-skirted PCR v-bottom plates. Arrange the samples vertically by column (i.e. A1, B1, C1, etc.)
![]()
Ship samples directly to our facility. Use the shipping address listed on the order receipt.
What can I do if my samples do not meet the starting material requirements?
At GENEWIZ, we know that samples are valuable and not always perfect. We're here to help find solutions for working with less-than-ideal samples. Please reach out to us by submitting an inquiry, and we’ll work with you to meet your project needs.
How do I request a quote?
For a quick tutorial, watch the video below.
How do I confirm a quote?
For a quick tutorial, watch the video below.
How can I monitor the progress of my project?
The status of your order, including an estimated date of delivery, can be viewed anytime through your GENEWIZ account. Visit the Order Summary page of your project to find the current order status.
How can I send my samples to GENEWIZ?
Option #1: Ship samples directly to our facility. Use the shipping address listed on your order receipt.
Option #2: For double-stranded DNA samples, submit samples into a local GENEWIZ dropbox which are conveniently located throughout the US, Europe and United Kingdom. You can easily locate a dropbox near you by logging into your account and choosing “Select from my dropboxes" during the order checkout process. Place your order receipt and samples in a Ziploc bag packaged according to sample submission guidelines, and place in dropbox for pickup. If you do not have a dropbox near you, please request a new dropbox location here.
- Note: Not recommended for RNA and primary sample types.
What are the cutoff times for my dropbox pickup?
During checkout, consult the Order Summary page to find out the daily cutoff times for your selected dropbox (see example below).

If you miss the dropbox deadline, feel free to ship samples directly to us.