Single-Cell ATAC-Seq Frequently Asked Questions
General Questions
What is single-cell ATAC-Seq?
Single-cell assay for transposase-accessible chromatin sequencing (scATAC-Seq) allows researchers to analyze chromatin accessibility in thousands of individual cells per sample. This level of throughput analysis provides insights into cell types and states, and provides a deeper understanding of gene regulatory mechanisms.
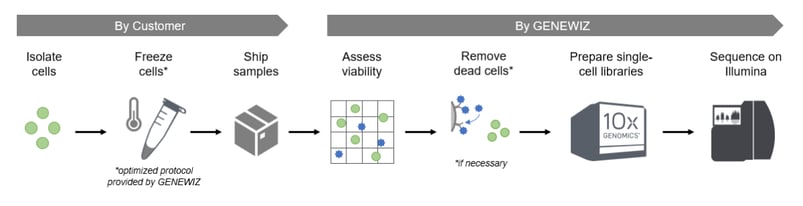
The workflow for scATAC-Seq is outlined below. Isolated cells are frozen by the customer according to GENEWIZ from Azenta's cryopreservation protocol and shipped overnight. At GENEWIZ, cell viability is assessed, and dead cell removal may be performed if necessary (see below for more information). Next, cells are processed by the 10x Genomics® Chromium™ Controller to create single-cell libraries. The libraries are then pooled and sequenced on the Illumina® platform.
How does scATAC-Seq differ from bulk ATAC-Seq?
ATAC-Seq is a bulk sequencing technology used to assess genome-wide chromatin accessibility. One caveat with bulk ATAC-Seq is that the chromatin signal is averaged for all cells in a sample, limiting insights into low-abundance phenotypes or rare populations of cells. scATAC-Seq enables researchers to interrogate the chromatin landscape of hundreds to thousands of individual cells at the single-cell level.
What is GENEWIZ's sample processing workflow?
Our workflow is illustrated here. To ensure the highest-quality data, viability and cell counts are measured immediately upon sample receipt, as cells must be processed on the Chromium Controller as soon as possible. As such, we developed a predefined workflow for when samples do not pass the initial QC for viability or cell count.
How do dead cells impact data quality?
Low cell viability causes unreliable cell recovery in the Chromium Controller and typically leads to missed sequencing targets and suboptimal results. Furthermore, ambient DNA fragments released by dead or apoptotic cells increase background noise, compromising data quality. To maximize the amount of useful sequencing data in single-cell projects, the presence of dead cells must be minimized. We recommend that cell viability exceeds 90%, with a minimum of 70%. Samples with less than 70% viability may undergo dead cell removal (DCR). A minimum of 106 cells and no previous treatment with magnetic beads is required for DCR. See our workflow for more information.
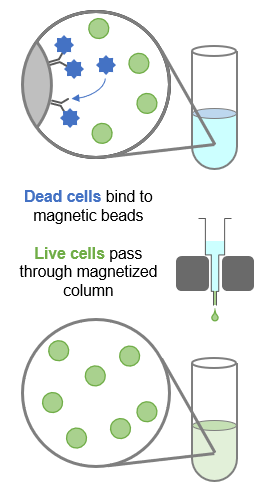
How does dead cell removal work?
Dead cell removal (DCR) uses magnetic beads conjugated to antibodies against annexin V, a marker for dead and apoptotic cells (see below). The beads capture dead or dying cells, enriching the sample for live cells.
How do I contact GENEWIZ for a technical consultation about my project?
GENEWIZ's NGS team is composed of Ph.D. scientists who can help you optimize your project design and provide consultation. Contact us here.
What is the price for scATAC-Seq?
Please submit a quote request to receive accurate pricing information, as cost varies depending on the details of the project.
Sample Preparation
How should I prepare and send my samples?
Sample Type: Cryopreserved single-cell suspension according to GENEWIZ supplied protocol, submitted in duplicate
Recommended Amount: >1M cells in 1 mL
Minimum Amount: 100,000 cells in 500 µL (Note: 1M cells are required for dead cell removal)
Buffer: Cryopreservation media (e.g. cell culture media supplemented with 20% FBS and 10% DMSO)
Review our single-cell ATAC-Seq best practices checklist for sample submission.
Note: GENEWIZ has developed a proprietary cryopreservation protocol to aid shipment of samples. Learn more about our protocol.
What happens if my samples do not meet GENEWIZ's starting material requirements?
Please contact our NGS team.
Sequencing & Data Analysis
Which platform and read length will be used for my project?
We use the 10x Genomics Chromium Controller coupled with Illumina NGS sequencing. The Chromium Controller uses a microfluidic chip that partitions cells across tens of thousands of Gel Beads-In-Emulsion (GEMs). This combination allows researchers to bioinformatically identify reads from hundreds to thousands of single cells in high-throughput fashion. 10x scATAC libraries use paired-end sequencing and dual indexing with the following configuration:
- Read 1: 50 cycles
- i7 Index: 8 cycles
- i5 Index: 16 cycles
- Read 2: 50 cycles
How many cells can be sequenced per sample on the Chromium Controller?
Revealing the full diversity of epigenetic regulation across a tissue or cell population often requires analysis of hundreds to thousands of viable cells. With our single-cell workflows, researchers have the flexibility to specify the number of cells to be sequenced, from 500 to 10,000 cells per sample. We typically recommend targeting 3,000 cells per sample for most experiments, but up to 10,000 cells for highly diverse populations.
How many reads do I need for my experiment?
The number of reads required depends upon genome size, number of known genes, cell type, and chromatin status of a sample. For most applications, 50,000 read pairs is the minimum read depth for scATAC-Seq (25,000 read 1; 25,000 read 2). We recommend 75,000 total reads per cell to maximize the identification of open chromatin regions in each cell.
What type of data analysis is available?
Complimentary scATAC-Seq analysis includes:
- Normalization of input runs to same median fragments per cell (sensitivity)
- Detection of accessible chromatin peaks
- Count matrix generation for peaks and transcription factors for the aggregate data
- Dimensionality reduction
- Cell clustering
- Cluster differential accessibility
How is data delivered?
GENEWIZ will recommend the best data delivery option based on the platform and project details. Data delivery options include:
- Secure File Transfer Protocol (sFTP)
- Customer cloud account
- Hard drive (additional charges may apply)
How will my data be delivered?
If your results are being shipped to you via hard drive, you can track its shipment using the tracking number provided to you in the delivery email sent from GENEWIZ.
If your results are being delivered via another data delivery method, such as AWS S3, Aspera, or other cloud-based methods, please refer to the delivery email sent from GENEWIZ for detailed information on the transfer, and consult with your IT department to ensure compliance with your institution’s policies.
If your results are being sent via a secure File Transfer Protocol (sFTP), please refer to GENEWIZ's sFTP Data Download Guide for instructions on how to download your data and troubleshooting tips.
Order & Processing
How do I request a quote?
Please fill out a quote request and our NGS team will follow up with you shortly.
What happens after I submit a quote request form?
Our NGS team will review your inquiry, follow up with a quote, and guide you through the next steps.
How do I confirm a quote?
For a quick tutorial, watch the video below.
How do I submit the GENEWIZ Sample Submission Form to provide detailed information about my samples?
Upon project confirmation, our NGS team will share our Sample Submission Form. Please complete the Form and send it back to NGS@azenta.com. Our team will update the details of the project on your behalf.
How can I monitor the progress of my project?
The status of your order, including an estimated date of delivery, can be viewed anytime through your GENEWIZ account. Visit the Order Summary page of your project to find the current order status. Watch the video below for a brief tutorial.
How long does GENEWIZ hold samples?
We hold samples for 1 year after project completion. Please contact us if you would like samples retained for a longer period or shipped back to you.
Have a specific question?
Contact Us | Phone 1-877-GENEWIZ (436-3949), ext. 1