Exome Sequencing Frequently Asked Questions
General Questions
1. What is exome sequencing?
Exome sequencing is the targeted enrichment and subsequent sequencing of the whole exome. Exome sequencing is potentially the most powerful tool available to the research community for the identification of genetic variations associated with a phenotype, such as a disease.
2. What is the advantage of exome sequencing compared to whole genome sequencing?
Whole genome sequencing requires an extremely high amount of sequencing throughput to generate a moderate depth of coverage. The data generated, while comprehensive, does not allow detection of mutations with as much sensitivity as a targeted approach. Exome sequencing is the most cost-effective and efficient solution.
A large portion of relevant mutations occur in the exome. In fact, the exome contains as many as 85% of disease-related mutations. Covering less than 2% of the whole genome, exome sequencing requires only 1/50th of the sequencing throughput to generate the same depth of coverage. This approach provides flexible experimental options:
- Maintain the same depth of coverage and multiplex more samples into the same sequencing lane, significantly decreasing total project cost
- Increase the depth of coverage to facilitate the detection of rare, low-frequency mutations
3. What species can be analyzed for exome sequencing?
Human and mouse exome sequencing are offered as standard services. However, we can procure commercially available exome kits for other species upon request.
4. What starting materials are accepted?
We routinely accept genomic DNA and have extensive experience performing extractions from a wide variety of materials, including cell pellets, fresh frozen tissue, blood, and FFPE slides. For more information, check out our Sample Submission Guidelines for research-use-only or clinical samples.
5. What exome kit do you use?
For human samples, we use Twist Human Comprehensive Exome Panel chemistry. However, other kits can be procured upon request.
6. How much coverage do I need?
The answer depends on the goals of your sequencing project. Several recommendations are shown below. Please feel free to contact us to discuss the right amount for your project.
- Germline/frequent variants: 50-100x (~5-10 Gb)
- Somatic/rare variants: ≥200x (20+ Gb)
- Tumor vs Normal: ≥200x (20+ Gb) tumor, ≥100x (10+ Gb) normal
- Population studies: 50-100x (~5-10 Gb)
7. Do you offer exome sequencing for clinical research?
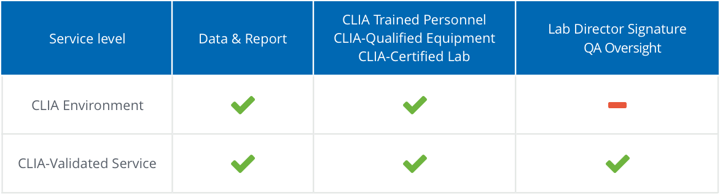
Yes! We offer two service levels for clinical whole exome sequencing: CLIA Environment and CLIA-Validated. Both are performed in a CLIA-certified lab by CLIA-trained personnel on CLIA-qualified equipment. The only difference is that CLIA-Validated Service includes a Lab Director Signature and enhanced QA Oversight whereas CLIA Environment does not. In addition, the CLIA Environment service level allows greater flexibility in experimental design, such as custom starting materials, and offers a range of sequencing coverage options. CLIA-Validated is recommended if results are used for diagnostic purposes. CLIA Environment is ideal for data that will not be reported back to patients or used for medical decision-making.
8. What bioinformatic analysis is available for exome sequencing projects?
We provide raw data as FASTQ files for all projects. We also offer mapping and variant calling through BAM and VCF files, respectively. CNV discovery is available for our research-use-only services. Please note that no variant annotation or interpretation is offered at this time for our clinical services.
9. Why choose GENEWIZ for exome sequencing projects?
We have proven our steadfast commitment to superior service throughout the years, becoming established as a global leader in DNA sequencing and genomics. Further, we have extensive experience and expertise in next generation sequencing techniques, including exome sequencing. An expert Project Manager can assist you in determining specifications for optimizing your project.
10. How is data delivered?
Yes, we can recommend the best data delivery option based on the platform and project details. Options include:
- Secure File Transfer Protocol (SFTP)
- Customer cloud account
- Hard drive (additional charges may apply)
11. How will my data be delivered?
If your results are being shipped to you via hard drive, you can track its shipment using the tracking number provided to you in the delivery email sent from GENEWIZ.
If your results are being delivered via another data delivery method, such as AWS S3, Aspera, or other cloud-based methods, please refer to the delivery email sent from GENEWIZ for detailed information on the transfer, and consult with your IT department to ensure compliance with your institution’s policies.
If your results are being sent via a secure File Transfer Protocol (sFTP), please refer to GENEWIZ’s sFTP Data Download Guide for instructions on how to download your data and troubleshooting tips.
11. What is the price for exome sequencing?
Please submit a quote request to receive accurate pricing information, as the cost depends on the details of the project.
12. What service packages are available?
We have three service packages available for human research-use-only exome sequencing. They differ in price and turnaround time:
- Value is the most economical: ~10% less in cost and ~2 weeks longer than Preferred
- Preferred offers a balance of speed and cost, leveraging our optimized processes for industry-leading turnaround time at an affordable price
- Express is the fastest: ~1 week faster and ~20% more in cost than Preferred
Furthermore, Preferred and Express options include guaranteed turnaround time and dedicated project managers. The package type can be revised after receiving your initial quote. For more information, visit our selection guide.
Note: Service packages are not available for all services, projects, or institutions. Clinical exome sequencing is excluded.
Sample Preparation
13. How should I prepare and send my samples?
View our Sample Submission Guidelines for instructions on preparing and sending your research-use-only or clinical samples for exome sequencing. Ship samples directly to our facility. Use the shipping address listed on the order receipt.
14. What happens if my samples do not meet your starting material requirements?
Please contact us.
Order & Processing
15. How do I request a quote?
For research-use-only projects, log in to our online system, navigate to the Next Generation Sequencing section, and select the Exome Sequencing icon.
For clinical projects, please request a quote here.
16. How do I confirm a quote?
For a quick tutorial, watch the video below.
17. How do I submit the GENEWIZ sample form (to provide detailed information about my samples)?
For a quick tutorial, watch the video below.
18. How can I monitor the progress of my project?
The status of your order, including an estimated date of delivery, can be viewed anytime through your GENEWIZ account. Visit the Order Summary page of your project to find the current order status.
19. How long do you hold samples?
We hold samples for 1 year after project completion. Please contact us if you would like samples retained for longer or shipped back to you.
Have a specific question?
Contact Us | Phone 1-877-GENEWIZ (436-3949), ext. 1