Sequencing Only Frequently Asked Questions
General Questions
1. What type of premade libraries can I submit?
You can submit any Illumina®-compatible library for sequencing, such as libraries from genomic DNA for whole genome sequencing or RNA for transcriptome sequencing.
2. How do I prepare my libraries for sequencing?
You can use any Illumina-compatible library preparation kit.
3. What sequencing platforms and configurations are available?
See below. Please contact us if your project requires a different platform or configuration.
4. What does it mean to be a standard Illumina-compatible library?
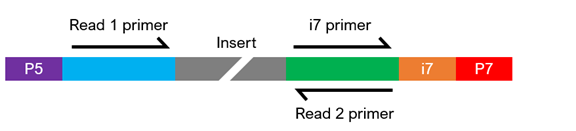
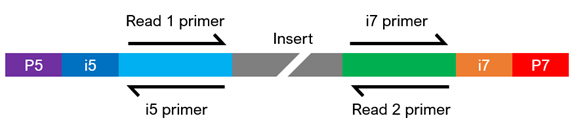
Illumina-compatible libraries require four major components in their adapter sequences, found at the ends of each fragment, for paired-end sequencing:
- Flow cell binding sites. Known as P5 or P7, they facilitate clustering of the DNA strands on the flow cell.
- Sequencing primer binding sites. Labeled as Read 1 and Read 2, they are necessary to initiate sequencing at both ends of the fragment.
- Index sequences. Often called i5 and i7, they act as barcodes to allow multiplexing (pooling) of samples. Libraries can be either single index (having one index sequencing) or dual index (having two index sequences), and each library to be pooled requires at least one unique index.
- Index primer binding site(s). These are required to read the index sequences.
The composition of the adapter sequences depends on whether the library is single or dual indexed (see figure below). If any of the components are missing, heavily modified, or in the wrong configuration, sequencing may fail. If you have any questions regarding your library configuration, please contact us.
Single-Indexed Sequencing
Dual-Indexed Sequencing
5. Can you spike PhiX into my sequencing run?
Yes, please instruct our team what percentage of PhiX spike-in to use, and we’ll accommodate the request free of charge. We typically recommend ~10% PhiX spike-in for projects with low diversity libraries.
6. Can you perform data analysis for my sequencing-only project?
Yes, please indicate this on the quote request form.
7. What are your starting material requirements for premade libraries?
We request that each library or library pool be ≥15 nM, ≥20 µL, in water or EB buffer. If you cannot meet these requirements, please contact us for further details.
8. How do I ship my premade libraries to you?
We request that you ship your libraries overnight on dry ice to our laboratory in Leipzig, Germany (address located in the quotation).
9. What QC is performed on the premade libraries?
We will run your libraries on the Agilent2100 Bioanalyzer to check the insert size and perform final quantification via qPCR prior to loading.
10. Can GENEWIZ de-multiplex my data?
We ask that you provide the index sequences in the Sample Submission Form prior to your sequencing run. GENEWIZ will provide the de-multiplexing free of charge. Alternatively, if no index sequences are provided, we will deliver the raw data and you can perform the de-multiplexing.
11. Can GENEWIZ perform de-multiplexing of “in-line” barcodes in sequencing reads?
Yes, we offer de-multiplexing of in-line barcodes in sequencing reads (not index reads). Please contact us for further details. Additional charges may apply.
12. Do you accept sequencing libraries with custom sequencing or index primers?
Yes, please contact us for further details.
13. How do I contact GENEWIZ for a technical consultation about my project?
GENEWIZ's NGS Team is composed of Ph.D. scientists who can help you optimize your project design and provide consultation. Contact the team via email at ngs.europe@genewiz.com or call us at +49 (0)341 520 122-41.
14. Can you recommend the best data delivery option?
Yes, we can recommend the best data delivery option based on the platform and project details. Options include:
- Secure File Transfer Protocol (sFTP)
- Customer cloud account
- Hard drive (additional charges may apply)
15. How will my data be delivered?
If your results are being shipped to you via hard drive, you can track its shipment using the tracking number provided to you in the delivery email sent from GENEWIZ.
If your results are being delivered via another data delivery method, such as AWS S3, Aspera, or other cloud-based methods, please refer to the delivery email sent from GENEWIZ for detailed information on the transfer, and consult with your IT department to ensure compliance with your institution’s policies.
If your results are being sent via a secure File Transfer Protocol (sFTP), please refer to GENEWIZ’s sFTP Data Download Guide for instructions on how to download your data and troubleshooting tips.
16. What is the price for NGS?
Please submit a quote request to receiving accurate pricing information, as the cost depends on the details of the project.
Sample Preparation
17. How should I prepare and send my samples?
- Sample Type: Premade Sequencing Library
- Recommended Amount: >15 nM, >20 µL
- Buffer: Water, EB, or low TE (<0.1 mM EDTA)
- Shipment Method: Dry ice
18. What happens if my samples do not meet your starting material requirements?
Please contact us via email at ngs.europe@genewiz.com or call us at +49 (0)341 520 122-41.
Order & Processing
19. How do I request a quote?
For European customers, please use our online form.
20. How long do you hold samples?
We hold samples for 1 year after project completion. Please contact us if you would like samples retained for a longer period of time or shipped back to you.
Have a specific question?
Contact Us | Phone 1-877-GENEWIZ (436-3949), ext. 1